H-NS is a bacterial transposon capture protein

Please read the article at:
https://doi.org/10.1101/2024.02.16.580519
Disclaimer: Below is an interpretation of the article thus do not necessarily represent the views of the authors.
Introduction
Histone-like nucleoid structuring proteins (H-NS) are regulators of expression in γ-proteobacteria which bind to DNA and inhibit transcription via xenogeneic silencing. Their preferential binding to AT-rich regions helps silence metabolically disruptive externally acquired DNA. These regions often contain promoters which can direct more RNA polymerases to their respective genes, thus diverting resources away from housekeeping genes, reducing fitness. This, among other functions, make H-NS a protective force for bacterial genomes.
Acinetobacter baumannii is the WHO top priority bacterial multi-drug resistant pathogen and is notorious for its ability to acquire and incorporate antimicrobial resistance genes (ARGs) into its chromosome. This is partly attributed to existing mobile genetic elements such as Insertion Sequences (IS). These are the simplest forms of transposons which contain only the transposase gene, capable of excising the IS and transposing it onto another location.
Results
Cooper et. al. noticed unusual grey colonies among their usually translucent A. baumannii AB5075 cultures. AB5075 normally contains two copies of ISAba13, however, sequencing revealed another ISAba13 inserted within the K-locus of the grey variants, a site containing capsule forming genes. This insertion resulted in the downregulation of K-locus genes and resulted in biofilm formation, reduced motility and susceptibility to human serum. Interestingly, the most downregulated gene was pilA, 0.4Mb away from the K-locus, suggesting collateral effects of the insertion.
Using a technique coined native Tn-seq, the transposition sites of ISAba13 were mapped. Transposition is rare, therefore most genomes only had the original two copies. Among those with transpositions, 25% inserted within non-coding regions. Most common insertion sites were AT-rich and not biased based on gene direction. Deletion of the H-NS gene resulted in more even distribution of insertions but no difference in total frequency.
H-NS does not only affect chromosomal DNA. Expression of H-NS resulted in a transposition bias into 2 out of 3 plasmids and 7 out of 11 prophages in AB5075, most notably within restriction modification systems. This may further reduce genetic mobility.
Genes bound by H-NS appear to be non-housekeeping and H-NS deletion removes this bias therefore cancelling its protective effect on housekeeping genes. Insertion into these would disable critical functions likely resulting in cell death.
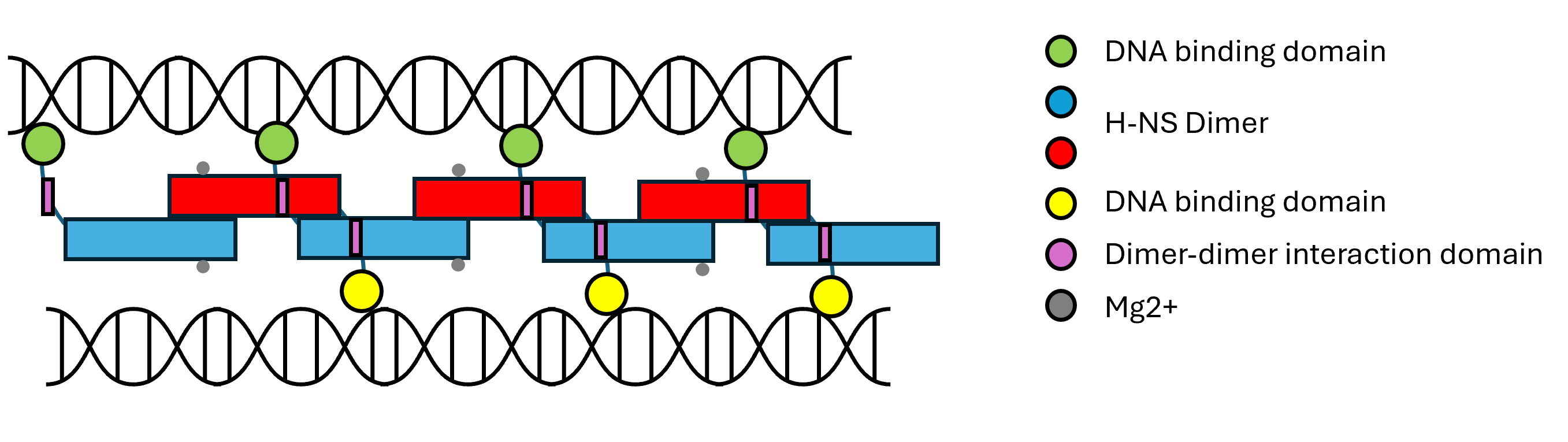
H-NS forms dimers which further polymerises with other dimers to form extended nucleoproteins. These polymerisation sites are potent DNA bridging inhibitors normally inactivated when polymerised and in the presence of magnesium ions. Cooper et. al. synthesised this 39 amino acid region (H-NS-39), demonstrated their ability to inhibit bridging but not binding, and used them deduce how H-NS specifically promotes transposition.
They hypothesised that either H-NS facilitates bridging between bound DNA and existing chromosomal ISAba13 or H-NS captures excised ISAba13. If the former is true, interactions between ISAba13 and transposition hotspots would be elevated in the presence of H-NS. 3C-seq analysis, a technique used to analyse interactions between segments of DNA, showed that these ISAba13 regions were some of the lowest interacting. Presence of H-NS-39 also did not inhibit ISAba13's interactions with other loci. Their interactions were largely limited to approx. 10kb either side. Interactions outside transposition hotspots were "infrequent and uniform", in sharp contrast to Tn-seq data.
They concluded that H-NS preferentially binds to AT-rich regions of DNA and bridges with excised transposons. This close proximity facilitates their transposition through mechanisms not yet investigated.
Thoughts
Though I consider this very robust and convincing, further work remains to determine the extent of H-NS' importance in transposition. Similar results with other transposons should be demonstrated. More importantly, showing whether composite transposons can be captured or if H-NS only captures the first IS element from which more complex transposons can be built from.
This mechanism should also be investigated in other species. A. baumannii is known for having relatively low GC content, thus high AT%. In bacteria with lower chromosomal AT%, does H-NS facilitate more transposition onto potentially higher AT plasmids? Could this explain potential differences in transposon mobility into vectors between species?
Finally, globally disseminated lineages of A. baumannii are known to carry chromosomal resistance islands. These contribute to A. baumannii's reputation as a proficient acceptor of ARGs. Deciphering a historic or current role of H-NS in the formation and development of these islands would further strengthen the case that H-NS contributes to the persistent and dissemination of antimicrobial resistance.
